KEY TOOLS OF GENETIC ENGINEERING :-
- Genetic engineering or recombinant DNA technology can be accomplished by using following key tools.
(1) Restriction enzymes
(2) Cloning vectors
(3) Ligases
(4) Cometent host organism
Some of these tools are discusses in detail below
RESTRICTION ENZYMES
- Restriction enzymes or molecular scissors are used for cutting DNA.
(1) Two enzymes from E.coli that were responsible for restricting the growth of bacteriophage were isolated in 1963,one of them added methyl group to DNA and the other cut DNA into segments.
The later was called restriction endonuclease.
(2) The first restriction endonuclease Hindii was isolated by Smith Wilcox and Kelley (1968).They found that it always cut DNA molecules at a particular point by recognising a specific sequence of six base pairs known as recognition sequence.
(3) Besides Hindii, more than 900 restriction enzymes have been isolated, from over 230 strains of bacteria, each of which recognises different recognition sequences.
 |
| DNA |
(4) NAMING OF RESTRICTION ENZYMES
(a) The first letter is derived from the genus name and the next two letters from the species name of the prokaryotic cell from which enzymes are extracted.
(b) The Roman numbers afer name show the order in which the enzymes were isolated from the bacterial strain.
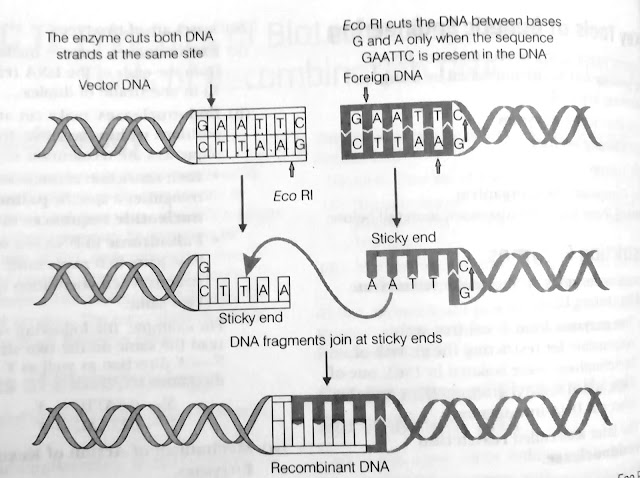
For example, Eco RI comes from the Escherichia coli RY13 and Eco RII comes from E.coli R 245,tc.
(5) Restriction enzymes belong to a class of enzymes called nucleases.
Nucleases are of two types
(a) Exonucleases remove nucleotides from the ends of the DNA (either 5' or 3') in one of duplex.
(b) Endonucleases make cut at specific position within the DNA. These specific sequences are recognition sequences.
* Each restriction Endonuclease recognises a specific Palindromic nucleotide sequences in the DNA.
* palindrome in DNA is a sequence of base pairs that reads same on the two strands when orientation of reading is kept sameb.
For example, the following sequences read the same on two strands in 5'--3' direction as well as 3'--5' direction.
5'----GAATTC----3'
3'----CTTAAG----5'
MECHANISMS OF ACTION OF RESTRICTION ENZYMES
(a) Restriction enzymes cut the strand of DNA a little away from the centre of the palindrome sites, but between the same two bases on the opposite strands.
(b) This leaves single - stranded portions at the ends.
(c) There are overhanging stretches called sticky ends on each strand. These are named so, because they form hydrogen bonds with their complementary cut counterparts.
(d) The stickiness of the ends faciliates the action of the enzyme DNA ligase.
(e) Restriction Endonuclease are used in genetic engineering to form recombinant molecules of DNA from different sources /genomes.
(f) These sticky ends are complementary to each other when cut by same restriction enzyme, therefore, can be joined together (end-to-end) using DNA ligases.
SEPARATION AND ISOLATION OF DNA FRAGMENTS :-
(1) The cutting of DNA by restriction endonucleases results in the fragments of DNA.
(2) the technique which separates DNA fragments based on their size is called gel electrophoresis.
(3) DNA fragments are negatively charged molecules. They can be separated by forcing them to move towards the anode under an electric field through a medium Matrix.
(4) the most common medium used as Agrose a natural polymer extracted form seaweeds.
(5) the DNA fragment separate resolve according to their size throw ceiling effect provided by the agrocel the smaller the fragments as the father it moves so small fragments of DNA travel more as compared to larger fragments.
(6) The separated DNA fragments can be visualised only after staining the DNA with a compound known as ethidium Bromide followed by exposure to UV radiation.
(7) The DNA fragments can be seen as bright Orange coloured bands. This separated bands are cut to from the agarose and extracted from the agarose gel, this is called elution.
(8) the purified DNA fragments can be used in constructing Recombinant DNA by joining them with cloning vectors.
Important topics of NEET 2020






![Nitin Vijay [NV] Sir Biography, Age, Salary, Wife Nad Much More](https://blogger.googleusercontent.com/img/a/AVvXsEh0YxSIuqtfzxRzl6NQdv-7Q58q8YLN7dPONEm8MoBjfTSF7wyhPsytw30B4uOHHiSnm70WUikIxcP5F4qiIUm5A8swamqBk_CGQqss0S4e7c02CQSraTSiiNKpvCVvG2v2qBrnFOBcVnhrqKBOvWvQlqQBRdz8HR-RADVHs4GIp9wbNnjJEfqF8mwO=w72-h72-p-k-no-nu)

0 Comments